What Are Codelists and How Are They Constructed?
- Posted:
- Written by:
- Categories:
This article is part of a series: Clinical Codes
- Prescribing Data: BNF Codes
- What is the dm+d? The NHS Dictionary of Medicines and Devices
- Difference between BNF, dm+d and SNOMED CT codes
- An Introduction to Clinical Codes and Terminology Systems
- What Are Codelists and How Are They Constructed?
This is one of a series of blog posts covering clinical codes and their use in OpenSAFELY research.
What are codelists?
Health data research, like the work done by the OpenSAFELY team, commonly relies on selecting patients with specific diagnoses, test results or medications prescribed through the use of clinical codes. There are often multiple clinical codes that can define a specific area of interest. A codelist is a collection of the relevant codes which define an area of interest. For example, in a study looking at patients with hypertension you would need a codelist to define the various types and subtypes of hypertension while excluding terms that are not relevant. We use the term codelist construction to define the process of creating a codelist. A simplified example is shown in the table below:
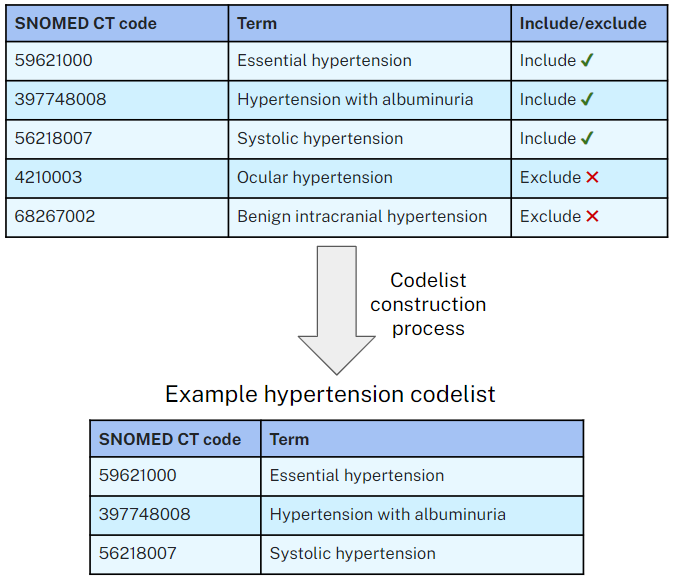
Example of inclusion/exclusion decisions when constructing a hypertension codelist
Terminology
We prefer to use the term codelist to describe a designed collection of clinical codes, however phrases such as code set, term set, reference set or refset might be used to refer to similar collections in different contexts. Codelists can also be used in conjunction with other logic rules to define cohorts of patients - this is referred to as a phenotype.
Codelist creation tools
We have developed OpenCodelists, a tool for creating and sharing codelists, which we use for the creation of all our codelists. OpenCodelists makes it quick and easy to search for and select clinical terms for inclusion or exclusion in the codelist. Full documentation on how to use OpenCodelists is available here.
Common pitfalls
Codelists can often be large and complex with some codelists made up of hundreds of individual clinical codes. Even a single incorrectly included or omitted code could potentially lead to vastly different results and present a misleading picture of what the underlying data shows. Some of the common pitfalls include:
- Including similar-sounding but unrelated codes: Using the above example of hypertension, there is a risk of inadvertently selecting codes for conditions unrelated to high blood pressure. For instance, ocular hypertension, which pertains to high fluid pressure within the eye, should be excluded to ensure accurate results.
- Omitting synonyms: Combinations of medical and non-medical terms can be used to describe a condition. It is important to be aware of and include common synonyms when creating a codelist. For example, when defining a codelist for sore throat, it is essential to include clinical codes which describe pharyngitis as well.
- Misunderstanding study intent: Selecting an appropriate codelist requires careful consideration of which patients are relevant to the research aims. For example; the decision to include or exclude gestational diabetes in a diabetes codelist, for instance, may vary depending on the specific study or context.
- Codes which can refer to both relevant and irrelevant cases: Some codes might be useful to improve sensitivity of a study but care needs to be taken to consider potential negative impact on specificity. For example, sore throat is a potential symptom of Group A Strep infection but is also a symptom of many other conditions, if including this in a study it is likely other codelists (for example antibiotics for Group A Strep treatment) would need to be used to maintain an appropriate level of specificity.
A carefully constructed, high quality codelist is therefore critical to ensuring reported outcomes in research are accurate and meaningful. This can be best achieved by following a standardised approach.
Existing standards and literature on codelist construction
Despite the widespread use of codelists within health data research there is no agreed ‘Gold-standard’ approach to their creation. To date, there has been a limited amount of published literature detailing methodology.
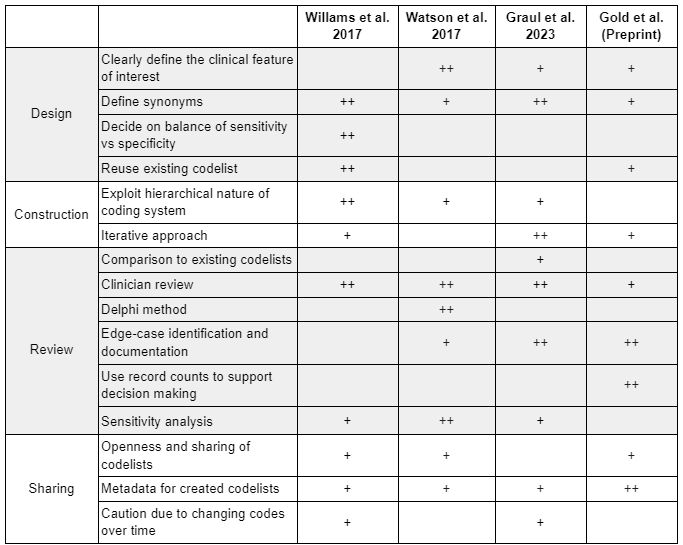
The most extensive piece of work on the topic to date, is the 2017 review article “Clinical code set engineering for reusing EHR data for research: A review” by Williams et al.1 which summarises approaches from 30 published methodological papers on clinical codelists. A number of commonalities in approaches were identified. The group then detailed a number of recommendations for codelist construction and management.
Use of the Delphi technique is a key part in the 2017 paper “Identifying clinical features in primary care electronic health record studies: methods for codelist development” Watson et al.2. They discuss detailed methodology of the approach followed by their group to create codelists. They have a careful approach to defining the purpose of the codelist and generating a list of synonyms to search for. Following generation of the codelist they utilise a Delphi review approach to ensure consensus between at least two clinicians on the final codelist.
Looking specifically at construction of medication codelists, the 2023 paper, “Determining prescriptions in electronic healthcare record (EHR) data: methods for development of standardized, reproducible drug codelists” by Graul et al.3 documents approaches which overlap with those also used for clinical codelists. They detail their full methodical approach, which has a particular focus on using a variety of search terms to capture a large range of potential codes followed by careful clinician review.
The preprinted work “Practices, norms, and aspirations regarding the construction, validation, and reuse of code sets in the analysis of real-world data” by Gold et al.4 used surveys and interviews of people who routinely create codelists to understand commonalities in approach. They found significant variability in formality of codelist construction approaches but more consistency in validation approaches. Their conclusion puts a particular emphasis on validation steps when constructing codelists and sharing of final codelists for reuse by other groups.
The below table summarises the key considerations identified across these papers (+), including an indication of the points which are given a particular emphasis (++).
In a future piece we will detail the Bennett Institute for Applied Data Science best practice guide for codelist construction.
References
-
Williams R, Kontopantelis E, Buchan I, Peek N. Clinical code set engineering for reusing EHR data for research: A review, Journal of Biomedical Informatics, Volume 70, 2017, Pages 1-13, https://doi.org/10.1016/j.jbi.2017.04.010 ↩︎
-
Watson J, Nicholson BD, Hamilton W, et al. Identifying clinical features in primary care electronic health record studies: methods for codelist development, BMJ Open 2017;7:e019637. https://doi.org/10.1136/bmjopen-2017-019637 ↩︎
-
Graul EL, Stone PW, Massen GM, Hatam S, Adamson A, Denaxas S et al. Determining prescriptions in electronic health care (EHR) data: methods for development of standardised, reproducible drug codelists JAMIA Open, Volume 6, Issue 3, October 2023 https://doi.org/10.1093/jamiaopen/ooad078 ↩︎
-
Gold S, Lehmann H, Schilling L, Lutters W. Practices, norms, and aspirations regarding the construction, validation, and reuse of code sets in the analysis of real-world data (preprint) medRxiv 2021.10.14.21264917; https://doi.org/10.1101/2021.10.14.21264917 ↩︎


